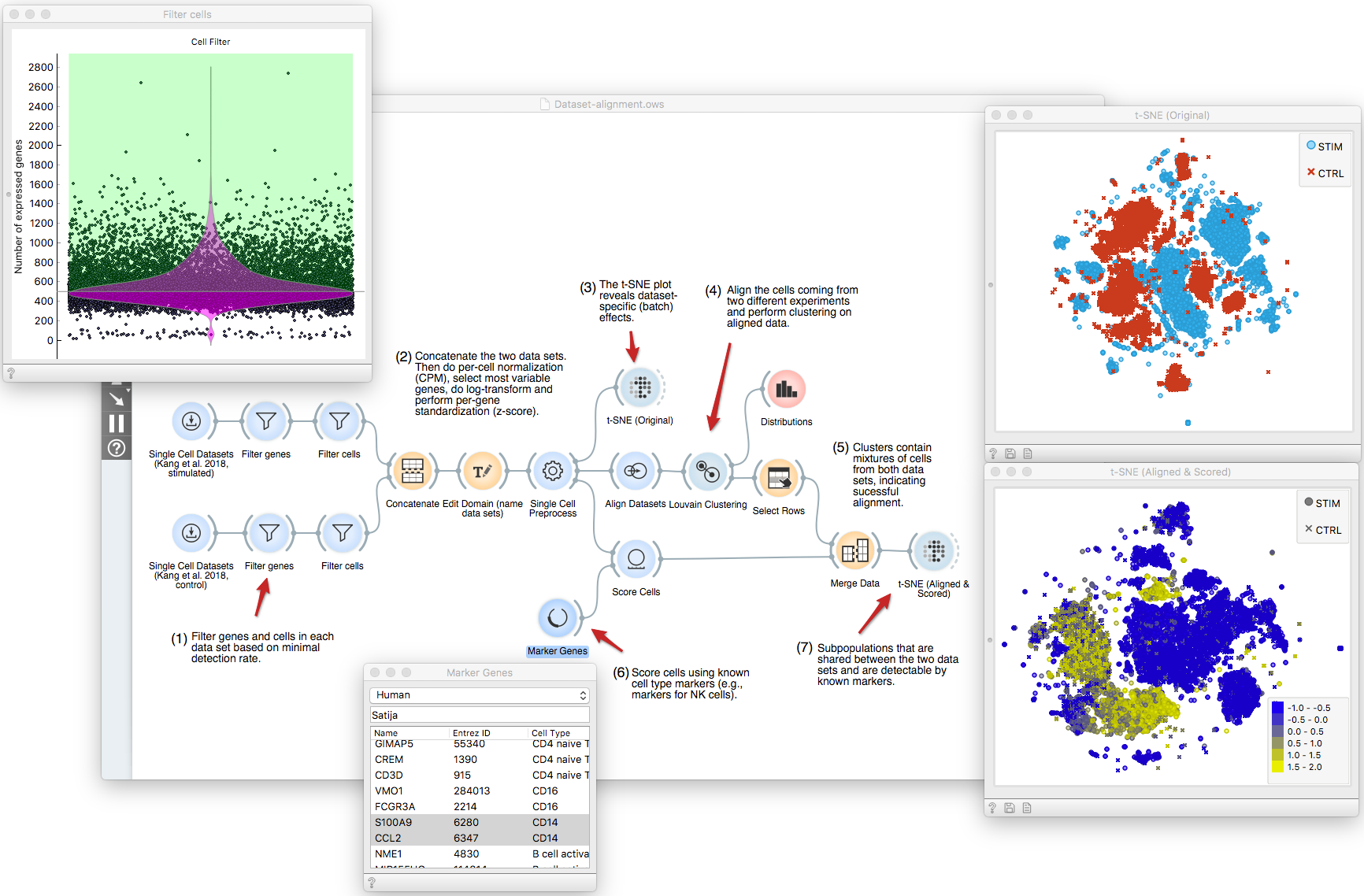
scOrange website is now live!
After a more than 15 year long venture with Orange Data Mining, the [Uni. of Ljubljana] Bioinformatics laboratory has embarked on a mission to extend the Orange canvas to single-cell analytics. Starting in the fall of 2017, scOrange (single-cell Orange) now includes an arsenal of widgets to support typical tasks with single-cell RNA sequencing datasets. These include:
- basic data manipulations (loading, annotation),
- filtering, normalization and other preprocessing,
- batch effects removal,
- clustering and cluster analysis,
- differential expression analysis,
- visualization.
All this comes with full power of Orange general purpose machine learning and data mining toolbox. Courtesy of Pavlin Poličar, We also have probably one of the most efficient implementations of the t-distributed Stochastic Neighbor Embedding (t-SNE), which is a go-to visualization method in these waters. It is able to project a million of cells into a 2D space in a couple of minutes by exploiting multi-core architectures and smart interpolation to approximate the distance matrix.
The main motivation to develop scOrange was a gap in interactive, open source applications for scRNA-seq, that do not require any coding. Despite being one year into coding and modelling, the general feeling is we just scratched the surface, and (we hope) many reproducible studies will be enabled by this product.
You are welcome to visit our website, which contains a handy blog with off-the-shelf tutorials, with videos to follow soon!