Reduce, reuse, recycle your t-SNE projections
Exploratory data analysis and modelling projects often start with a compact visualization of available data. Staples like principal component analysis (PCA) or multidimensional scaling (MDS) methods achieve this by finding a two dimensional summary of the data that preserves the main axes of variation. They reveal potential patterns and (if we are lucky) some structure in a multi-dimensional data set.
The t-distributed Stochastic neighbour Embedding (t-SNE) is a modern variation on these methods, that more closely resembles to how a human user might interpret such figures. Essentially, we only care that very similar points appear close together to a given point, while more distant points do not influence its positioning. This allows dramatically more appealing projections, that although harder to quantify, tend to produce clumped, grape-like clusters.
Few fields have used t-SNE to such extent as single-cell transcriptomics, where it provides a natural tool for revealing different cell types or states.
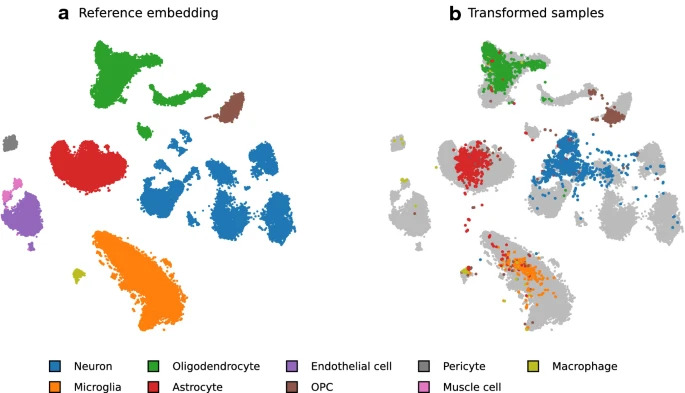
The non-linear and data-driven nature of t-SNE makes it hard to re-use and compare between different data sets. In our latest methods work, led by Pavlin G. Poličar, we develop a principled way to perform t-SNE with two different data set. One data set is termed the primary or the reference data set, where t-SNE is computed as usual. When bringing in a second data sets, we implement an efficient and parallel way to find the best position for each point in the reference projection.
Image the reference projection as a scaffold that establishes possible positions for each point. This way, large reference t-SNE projections of millions of cells can be used to make sense of new, smaller data sets that are generated!
Read more in Machine learning.