Microbial chemicals in early onset ulcerative colitis

Since the widespread use of antibiotics almost a century ago, acute bacterial infections have largely been largely superseded by chronic inflammation as a prominent cause of morbidity and mortality. Ulcerative colitis (UC) is a form of inflammation of the lower intestine, which may ultimately lead to colon cancers. The prevalence of these conditions in Western and Westernized population attributes disease risk to a mesh of genetic predispositions, environment or diet. While the exact triggers are still unknown, and may not be unique, gut bacteria are though to be primary suspects in these cases.
To get an early snapshot of the scene, our study led by dr. Melanie Schirmer examined pediatric UC patients, aged 4-17. The subjects have not undergone treatment, potentially promising a less obstructed view into onset of inflammation. Through investigation of paired stool and blood, we provide insights into bacterial communities and their chemical repertoire that characterizes ulcerative colitis at its earliest stages.
Various practices for exploring microbial compositions exist. We opted for de novo assembly, which takes bits and pieces of microbial DNA and reconstruct bacterial and even viral genomes, even if most these microbes have previously been unknown. As opposed to using known bacterial genome catalogs, this approach makes it harder to compare bacterial strains between studies, but has greater sensitivity due to thorough utilization of the collected material.
Inflammation at mucosal sites is characterized by low oxygen availability, prompting cells to switch from respiration to alternative metabolic pathways. These typically produce less energy per unit of sugar and result in accumulation of metabolic waste products such as lactic acid. Similar conditions arise during intensive exercise, where demands for oxygen skyrocket, potentially leading to hypoxia.
Patients with most severe inflammation harbored bacteria with special metabolic tricks up their collective sleeves. Most prominent of those was Veillonella parvula, which typically resides in the oral cavity but tends to relocate to the gut when inflammatory conditions favor its colonization. V. parvula is particularly suited to inflamed conditions, as it can utilize nitrate (NO3-) as an alternative to oxygen for respiration. Besides, lactic acid is one of its favorite nutrients, using it as a source of carbon. It is no surprise that Veillonella is also found in high abundances in marathon runners.
Subjects with Veillonella were also more likely to undergo colectomy, a surgery to treat the inflamed colon. However, the jury is still out on whether V. parvula is a mere sign of inflammation or contributes to disease exacerbation.
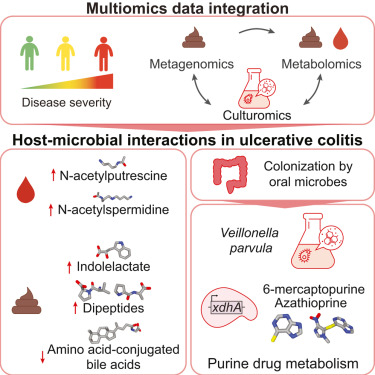
To address these questions, we cultured Veillonella in a dish and analyzed the molecules it makes in conditions similar to the inflamed gut. Besides lactic acid, it likes to consume dipeptides - debris from proteins that tend to be abundant in inflammation. We observed the emergence of numerous molecules associated with disease progression, including immunomodulatory metabolite indolelactic acid which may result from tryptophan-rich diet. We also detected lipids which can signal to sensory neurons (oleamide) and contribute to exercise motivation . Interestingly, we also found genes in Veillonella that help it meddle with thiopurines, molecules that are similar to immunosuppresive drugs used to treat inflammatory bowel diseases.
In summary, we seen how a microbe particularly tailored to live in inflamed guts characterizes disease states and produces bioactive molecules. Related nitrate addicts, such as Klebsiella pneumoniae, have previously been established as causative disease agents and are now targeted by phage therapies. As inflammatory bowel disease seem to be mosaic of environmental exposures identification of novel microbes and molecules aids diagnosis, therapeutic targets and recommendations.
This work was featured as a cover in Cell Host & Microbe, February 2024.